June 4, 2025
When images learn to walk: New method visualizes molecular movement
Simulations and machine learning enable the analysis of electron microscopy images in miliseconds
A new method makes it possible to better understand the motions of molecules which is important for their function. Researchers at FIAS, together with a team from the Flatiron Institute in New York (USA), have developed a technique to evaluate electron microscope images faster and more reliably with the help of physics-based simulations and machine learning. In the future this will help us gain deeper understanding on how the body works.
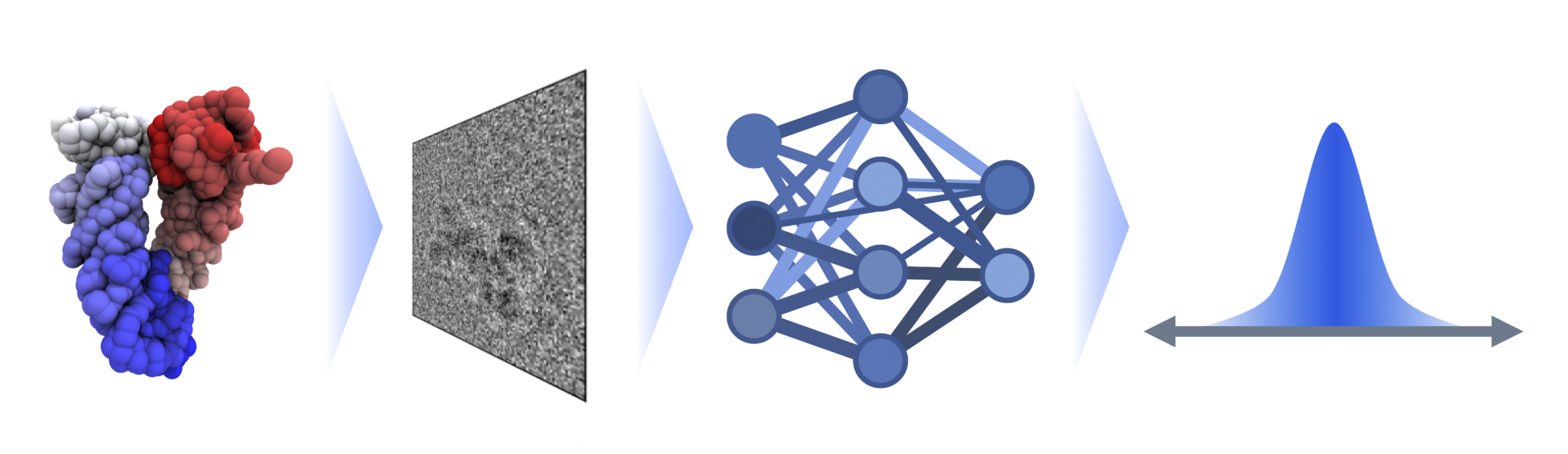
Biomolecules such as proteins or DNA are flexible and mobile - this is crucial for their function. However, images taken with modern electron microscopes only show static snapshots: in cryo-electron microscopy, the molecules are frozen and imaged in this static structure. “The individual images of these tiny structures are often noisy - and we don't know from which side the molecule is photographed,” says FIAS doctoral student Lars Dingeldein, describing the challenging analysis of such images. The group, led by FIAS Senior Fellow Roberto Covino, has therefore developed a method that calculates both the recording angle and the change in the molecules shape.
Dingeldein compares this to snapshots of a runner: “To understand the movement, you need many individual images”. The scientists therefore used biomolecules with a known movement profile to train their system: “We simulate the microscopy process and obtain a data set of artificial images of which we know the exact appearance,” says Dingeldein. This teaches the system to recognize the structure of a biomolecule based on the images. Machine learning is therefore based on known data - from the structure to the image - and can later use this experience to calculate the structure from images.
The 27-year-old biophysicist developed the algorithm with the help of the huge computing capacities. Once the system had been trained accordingly, the researchers applied the algorithm to experimental data sets from which the result was known. The structure and movement calculated using the new method matched the known data.
The new method simulation-based inference for cryo-EM (cryoSBI) developed by the FIAS researchers enables a significantly faster understanding of protein and gene structures: with conventional methods, it currently takes around 15 seconds to determine the shape of a protein for a single image. With cryoSBI, it only takes milliseconds. “Training is time-consuming, but the method is significantly more cost-effective afterwards,” says Dingeldein. The technology is now available to interested users. “It will enable us to understand biological systems faster and better,” Group Leader Covino is convinced: “For medical applications, for example, it is important to understand the movement and possible pathological disruption of biomolecules.” The team will continue to optimize the method and improve its application in the future.
Publication: Lars Dingeldein, David Silva-Sánchez, Luke Evans: Amortized template matching of molecular conformations from cryoelectron microscopy images using simulation-based inference, PNAS 122 (23) e2420158122, https://doi.org/10.1073/pnas.2420158122